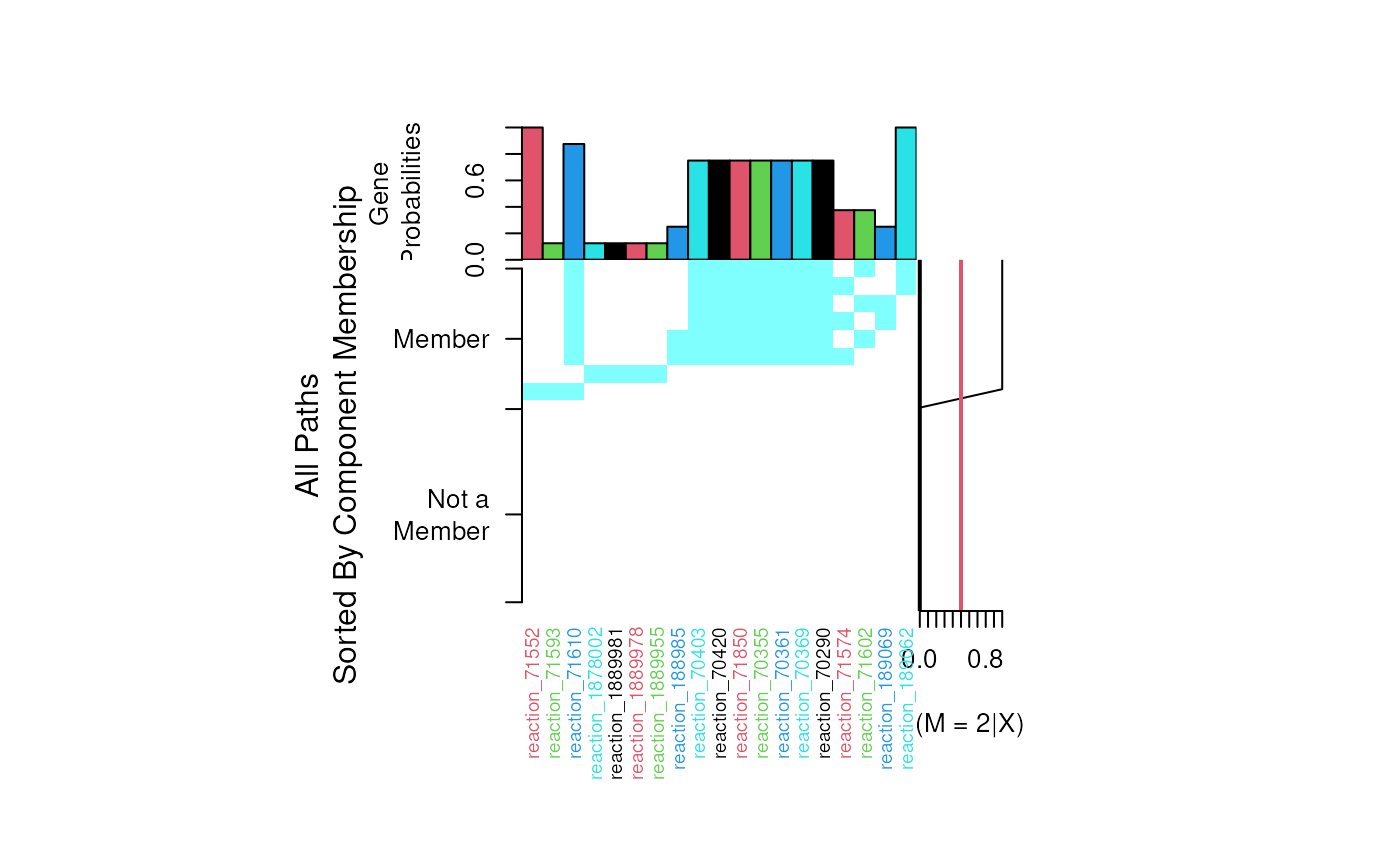Plots the structure of specified path found by pathCluster.
plotPathCluster(ybinpaths, clusters, m, tol = NULL)Arguments
- ybinpaths
The training paths computed by
pathsToBinary.- clusters
The pathway cluster model trained by
pathClusterorpathClassifier.- m
The path cluster to view.
- tol
A tolerance for 3M parameter
thetawhich is the probability for each edge within each cluster. If the tolerance is set all edges with athetabelow that tolerance will be removed from the plot.
Value
Produces a plot of the paths with the path probabilities and cluster membership probabilities.
- Center Plot
An image of all paths the training dataset. Rows are the paths and columns are the genes (features) included within each path.
- Right
The training set posterior probabilities for each path belonging to the current 3M component.
- Top Bar Plots
Theta, The 3M component probabilities - indicates the importance of each edge to a pathway.
See also
Other Path clustering & classification methods:
pathClassifier(),
pathCluster(),
pathsToBinary(),
plotClassifierROC(),
plotClusterMatrix(),
plotPathClassifier(),
predictPathClassifier(),
predictPathCluster()
Examples
## Prepare a weighted reaction network.
## Conver a metabolic network to a reaction network.
data(ex_sbml) # bipartite metabolic network of Carbohydrate metabolism.
rgraph <- makeReactionNetwork(ex_sbml, simplify=TRUE)
#> This graph was created by an old(er) igraph version.
#> ℹ Call `igraph::upgrade_graph()` on it to use with the current igraph version.
#> For now we convert it on the fly...
## Assign edge weights based on Affymetrix attributes and microarray dataset.
# Calculate Pearson's correlation.
data(ex_microarray) # Part of ALL dataset.
rgraph <- assignEdgeWeights(microarray = ex_microarray, graph = rgraph,
weight.method = "cor", use.attr="miriam.uniprot", bootstrap = FALSE)
#> 100 genes were present in the microarray, but not represented in the network.
#> 55 genes were couldn't be found in microarray.
#> Assigning edge weights.
## Get ranked paths using probabilistic shortest paths.
ranked.p <- pathRanker(rgraph, method="prob.shortest.path",
K=20, minPathSize=8)
#> Extracting the 20 most probable paths.
## Convert paths to binary matrix.
ybinpaths <- pathsToBinary(ranked.p)
p.cluster <- pathCluster(ybinpaths, M=2)
plotPathCluster(ybinpaths, p.cluster, m=2, tol=0.05)