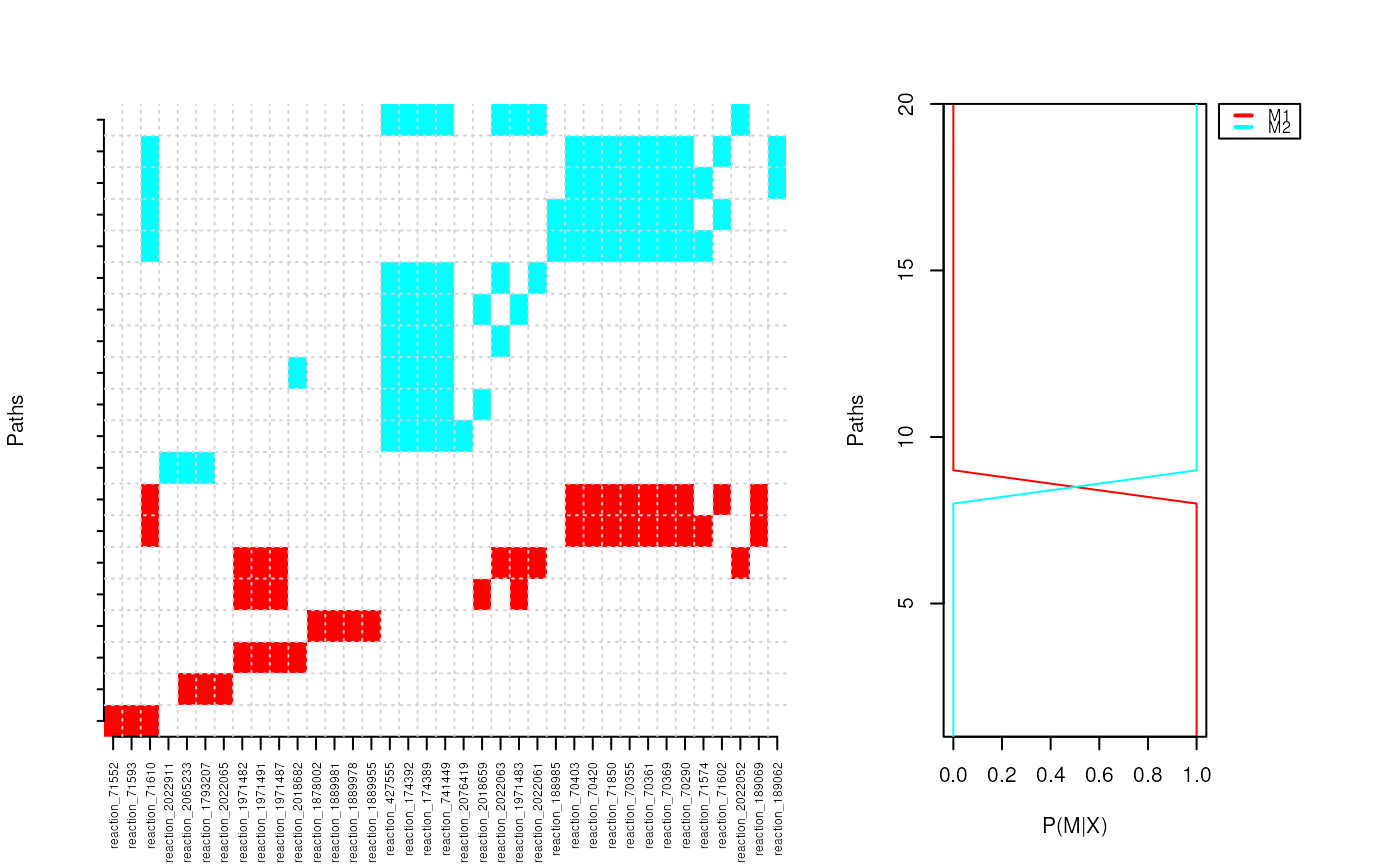
3M Markov mixture model for clustering pathways
pathCluster(ybinpaths, M, iter = 1000)Arguments
- ybinpaths
The training paths computed by
pathsToBinary.- M
The number of clusters.
- iter
The maximum number of EM iterations.
Value
A list with the following items:
- h
The posterior probabilities that each path belongs to each cluster.
- labels
The cluster membership labels.
- theta
The probabilities of each gene for each cluster.
- proportions
The mixing proportions of each path.
- likelihood
The likelihood convergence history.
- params
The specific parameters used.
References
Mamitsuka, H., Okuno, Y., and Yamaguchi, A. 2003. Mining biologically active patterns in metabolic pathways using microarray expression profiles. SIGKDD Explor. News l. 5, 2 (Dec. 2003), 113-121.
See also
Other Path clustering & classification methods:
pathClassifier(),
pathsToBinary(),
plotClassifierROC(),
plotClusterMatrix(),
plotPathClassifier(),
plotPathCluster(),
predictPathClassifier(),
predictPathCluster()
Examples
## Prepare a weighted reaction network.
## Conver a metabolic network to a reaction network.
data(ex_sbml) # bipartite metabolic network of Carbohydrate metabolism.
rgraph <- makeReactionNetwork(ex_sbml, simplify=TRUE)
#> This graph was created by an old(er) igraph version.
#> ℹ Call `igraph::upgrade_graph()` on it to use with the current igraph version.
#> For now we convert it on the fly...
## Assign edge weights based on Affymetrix attributes and microarray dataset.
# Calculate Pearson's correlation.
data(ex_microarray) # Part of ALL dataset.
rgraph <- assignEdgeWeights(microarray = ex_microarray, graph = rgraph,
weight.method = "cor", use.attr="miriam.uniprot", bootstrap = FALSE)
#> 100 genes were present in the microarray, but not represented in the network.
#> 55 genes were couldn't be found in microarray.
#> Assigning edge weights.
## Get ranked paths using probabilistic shortest paths.
ranked.p <- pathRanker(rgraph, method="prob.shortest.path",
K=20, minPathSize=8)
#> Extracting the 20 most probable paths.
## Convert paths to binary matrix.
ybinpaths <- pathsToBinary(ranked.p)
p.cluster <- pathCluster(ybinpaths, M=2)
plotClusters(ybinpaths, p.cluster)